Module 3: The Ecosystem Model
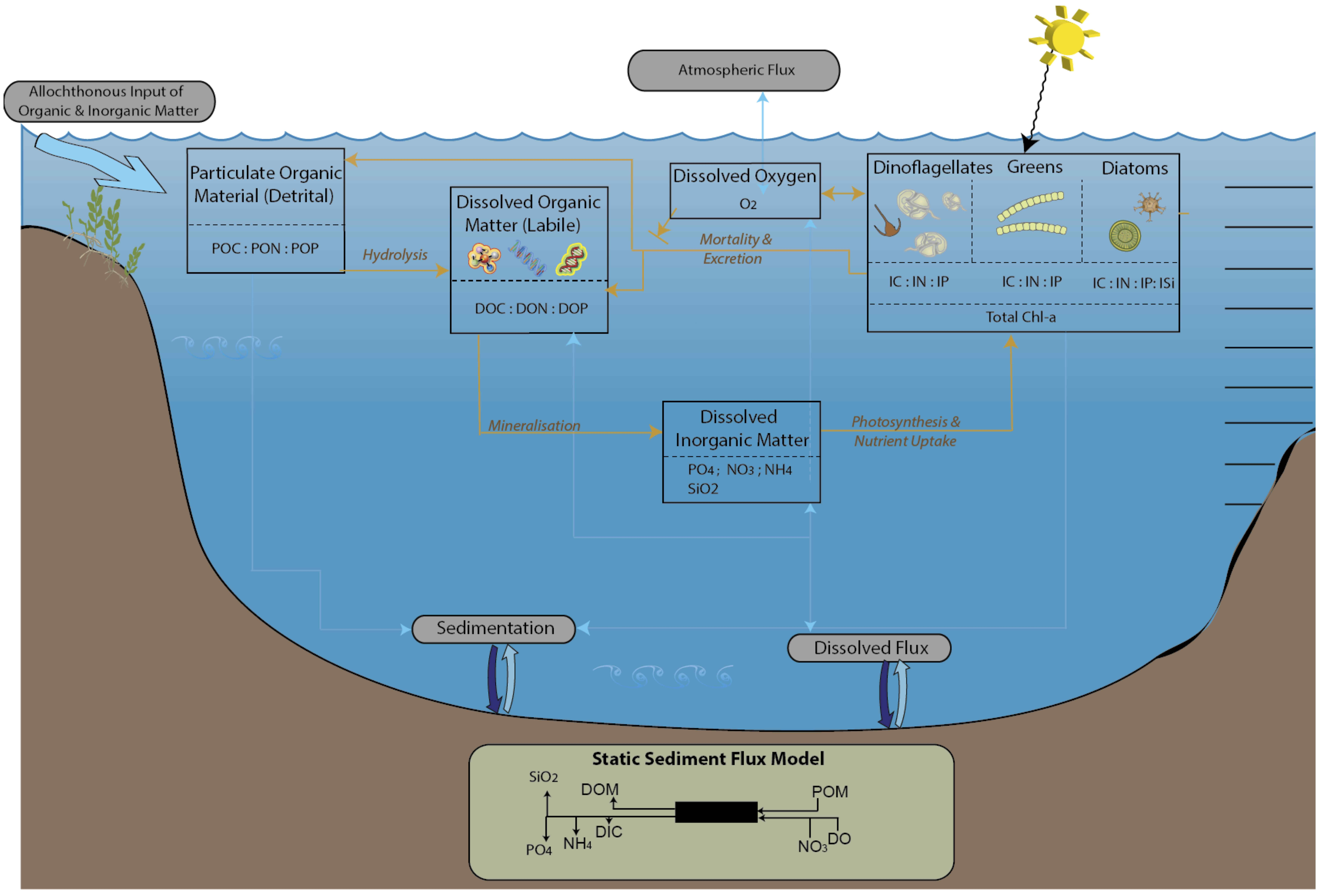
The Kinneret97 model simulation above has the ecological model aed2 enabled. It has been pre- configured to run the customizable AED modules. These are separate models, but have been set up to run side by side. They are configured using the text file aed2.nml.
The above simulation was only plotting temperature and salinity and these plots were configured in plots.nml. Open plots.nml to see how you can customise the graphs. There are many other variables we may be interested in plotting when we run the watter quality model such as oxygen, nutrients, and algae. A pre-configured file called plots_aed2.nml has been created. To use this file, rename it to plots.nml (you’ll first need to rename the existing plots.nml) and run the model. You should now see more outputs for the lake simulation.
In plots.nml the required variables are all listed, make sure the number of plots is high enough to include them all.
| Description | Variable Name |
|---|---|
| Passive tracer |
TRC_tr1
|
| Water age |
TRC_age
|
| Suspended solids |
NCS_ss1
|
| Dissolved oxygen |
OXY_oxy
|
| Dissolved inorganic carbon |
CAR_dic
|
| pH |
CAR_pH
|
| Methane (dissolved) |
CAR_ch4
|
| Methane (bubbles) |
CAR_ch4_bub
|
| Reactive Silica |
SIL_rsi
|
| Ammonium |
NIT_amm
|
| Nitrate/Nitrite |
NIT_nit
|
| Phosphate |
PHS_frp
|
| Particulate inorganic phosphorus |
PHS_frp_ads
|
| Dissolved organic carbon |
OGM_doc
|
| Particualte organic carbon |
OGM_poc
|
| Dissolved organic nitrogen |
OGM_don
|
| Particulate organic nitrogen |
OGM_pon
|
| Dissolved organic phosphorus |
OGM_dop
|
| Particulate organic phsophorus |
OGM_pop
|
| Green algae |
PHY_green
|
| Green algae |
PHY_green_IN
|
| Green algae |
PHY_green_IP
|
| Diatoms |
PHY_diatom
|
| Diatoms |
PHY_diatom_IN
|
| Diatoms |
PHY_diatom_IP
|
| Cyrptophytes |
PHY_crypto
|
| Cyrptophytes |
PHY_crypto_IN
|
| Cyrptophytes |
PHY_crypto_IP
|
| Zooplankton |
ZOO_zoo01
|
Let’s also now add water quality variables to the specific depth output files (i.e. WQ_5.csv). To do so we must edit the output section of the glm3.nml file, by adding the variables as extra columns to the .csv file that were configured above.
-
-
As an example to output water quality variables, let’s plot all the nitrogen variables. Change
csv_point_varsto includeNIT_amm,NIT_nit,OGM_don,OGM_pon,PHY_green_IN- these are all the variables that contribute to the total nitrogen (TN) pool. Now re-run the model (e.g. Windows users double clickingglm.batagain). Once it’s run openWQ_35.csvto see these new variables and create a stacked area plot to show how the different variables contribute to TN. -
Create a well formatted, interesting graph of variables of your choice (for example, you may like to consider all of the phytoplankton groups or create sums of these to generate a NPZD model) to see how they interact and change over time. If any variables are much bigger or smaller than the others, then use two y-axes or multiple plots.
-
- How would these concentrations change if the air temperature increased by 2°C? (You will need to edit the
met_hourly.csvfile). Make sure to save with correct date format, taking note these met measurements are recorded hourly, so YYYY-MM-DD hh:mm. A pre-made version of this file can be found in bcs which is calledmet_1997_2004_airT2.csv. To use this file, updatemeteo_fl = 'bcs/met_1997_2004.csv'in the&meteorologysection ofglm3.nml.
!-------------------------------------------------------------------------------
! meteorology
!-------------------------------------------------------------------------------
&meteorology
met_sw = .true.
lw_type = 'LW_IN'
rain_sw = .false.
atm_stab = 0
fetch_mode = 0
!rad_mode = 1
albedo_mode = 1
cloud_mode = 4
!-- BC file details
subdaily = .true.
meteo_fl = 'bcs/met_1997_2004.csv' !<------- Update this
wind_factor = 0.9
lw_factor = 1.0
lw_offset = 0.0
!-- Parameters
ce = 0.0013
ch = 0.0013
cd = 0.0013
!catchrain = .true.
!rain_threshold = 0.001
!runoff_coef = 0.0
/- What happens if we change the water clarity? Increase and decrease from the original value to see the response
Kw (range 0-1) in the glm3.nml file.