Module 8: Lake Water Quality Modelling
Module Resources
Download the GLM files for this module by clicking the download button in the tool bar .
Setting up GLM
Running GLM on MacOS
- Open the Terminal application on your Mac
- Use the
cd(change directory) command to navigate to the Kinneret97 folder - In Finder, go to the macGLM folder and right-click on the glm app, select Show Package Contents
- Enter the Contents folder then the MacOS folder, and locate the glm executable file
- Back in terminal, enter the file path for this executable file - you can do this by dragging/dropping the file into the terminal
- Leave a space after the file path and type
--xdisp - Press Enter and the model will open
Running GLM on Windows
On Windows, the model can be run using a batch-file glm.bat, which you can double click from in the Kinneret97 directory. The .bat file calls the GLM model from the current working directory.
If the model is not opening make sure you’re not working in a network directory (e.g. OneDrive) and the file path to the glm.bat file has zero spaces in it.
Running GLM in RStudio
If you want to use RStudio, open the R Project, then each of the R files: Get started.R, Change parameters.R and Plot.R. Run the lines in Get started.R - it will download all the R packages from the internet, as well as an old version of GLM.
To get the latest version of GLM, paste the folder winGLM into the place where R has saved its old version of winGLM, for example, H:/My Documents/R/win-library/3.4/GLMr/extbin/winGLM. After that you can drive the model and plot everything using the other two scripts.
Configuring Model Parameters
To configure the model, open the glm3.nml file in the Kinneret97 directory with a text editor. A chunk of this .nml* file is shown below. For example, if you wanted to change the maximum number of lake layers the model simulates, you would changemax_layers = 500`.
!------------------------------------------------------------------------------!
! !
! .----------------. .----------------. .----------------. !
! | .--------------. || .--------------. || .--------------. | !
! | | ______ | || | _____ | || | ____ ____ | | !
! | | .' ___ | | || | |_ _| | || ||_ \ / _|| | !
! | | / .' \_| | || | | | | || | | \/ | | | !
! | | | | ____ | || | | | _ | || | | |\ /| | | | !
! | | \ `.___] _| | || | _| |__/ | | || | _| |_\/_| |_ | | !
! | | `._____.' | || | |________| | || ||_____||_____|| | !
! | | | || | | || | | | !
! | '--------------' || '--------------' || '--------------' | !
! '----------------' '----------------' '----------------' !
! !
!-------------------------------------------------------------------------------
!-------------------------------------------------------------------------------
! general model setup
!-------------------------------------------------------------------------------
&glm_setup
sim_name = 'Example Kinneret GLM Simulation'
max_layers = 500
min_layer_vol = 0.025
min_layer_thick = 0.15
max_layer_thick = 1.50
density_model = 1
/Exercises
Lake Stratification
The simulation in the Kinneret97 directory is an example 40 m deep, monomictic lake that is used to supply drinking water and support an active fishery.
Before running the model, open the file glm3.nml in a text editor and inspect the model setup etc. This file is the main configuration file that drives the model. The file glm3.nml has several key sections in it related to the model domain and parameters, the sources of boundary condition data, and details of outputs. Scan the parameters, eg number of layers, light extinction coefficient, mixing coefficient etc.
Once the model has run, you can inspect the results. There are three ways to look at output:
- Via the time-depth contour plots that appear during the run (configured via
plots.nml) - Further information on the daily water and energy balance is in the output file
lake.csv - Time series values of selected state variables (e.g. temperature, salinity, etc…) can be requested for a
specific depth (depth above bottom) – this is setup in
&outputs(eg.WQ_5.csv)
- Draw a schematic picture of the lake, the relevant boundary conditions (both related to water fluxes and energy fluxes).
- Identify and open the boundary condition input files - these are csv files containing time-series of meteorological information, and inflow and outflow volumes. These files can be opened in Excel or a text-editor. Find where these files are called from within
glm.nml. - Plot the lake water balance – volume, water level and fluxes including inflows, outflows, rainfall and evaporation (refer to columns in
lake.csv, and copy the appropriate data into your own excel spreadsheet).
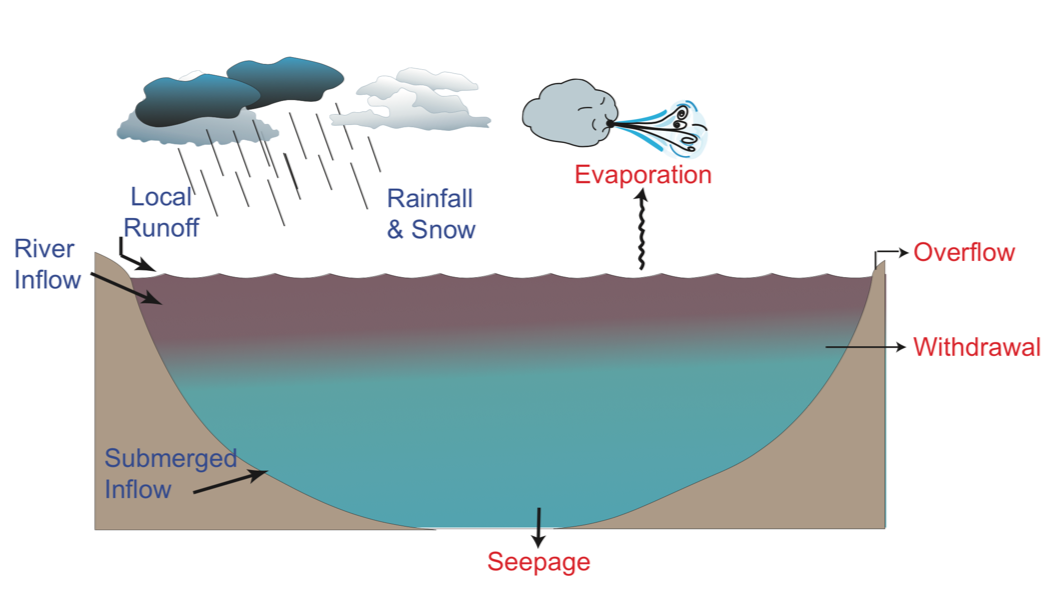
Figure 1: Schematic diagram of water balance components.
- Plot the surface heat fluxes such as shortwave, longwave, sensible heat latent heat (refer to columns in
lake.csv).
](images/09-module8/picture3.png)
Figure 2: Schematic diagram of surface heat fluxes impacting lake water temperature. For a description of the heat flux terms refer to the GLM paper
- Go to the
&outputsection ofglm.nmland customize the configuration to make two output files, one at 5 m from the bottom (ie. in the lake hypolimnion) and one at 35m from the bottom (ie. in the lake epilimnion). This create two.csvfiles (WQ_5.csvandWQ_35.csv) with conditions at these water depths. Plot the temperate of the two layers in excel. Identify the periods where the lake is stratfied and mixed.
!-------------------------------------------------------------------------------
! format for output and filename(s)
!-------------------------------------------------------------------------------
&output
out_dir = 'output'
out_fn = 'output'
nsave = 6 !This will output every 6 hours
! General summary file
csv_lake_fname = 'lake'
! Depth specific outputs
csv_point_nlevs = 2
csv_point_fname = 'WQ_'
csv_point_at = 5.,35.
csv_point_nvars = 4
csv_point_vars = 'temp','salt','PHS_frp','NIT_amm'
/- Assess how the degree of stratification changes as the initial water level of the lake is reduced by 20 m. To do this change the initial starting lake depth and profile details in glm.nml.
The Ecosystem Model
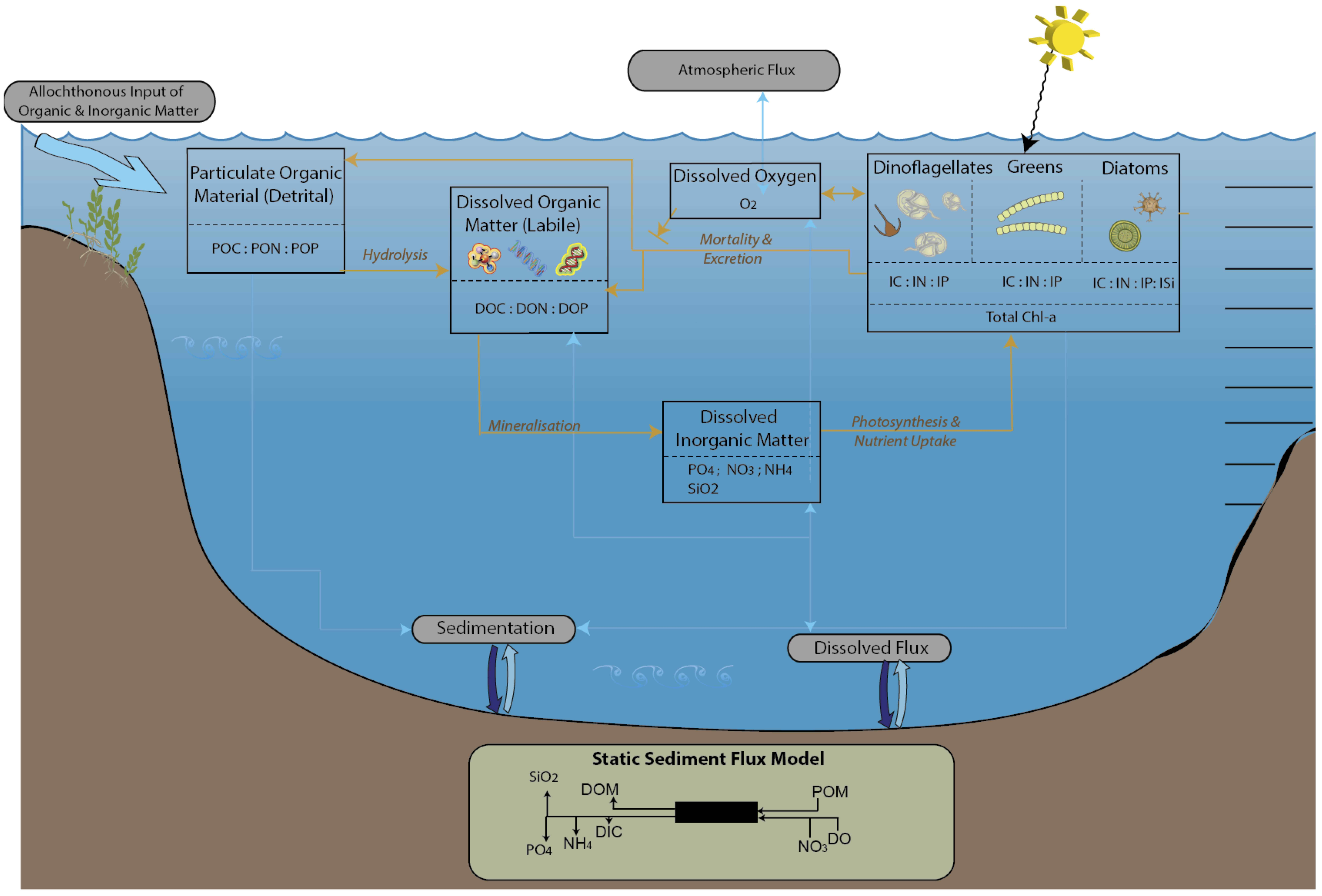
The Kinneret97 model simulation above has the ecological model aed2 enabled. It has been pre- configured to run the customizable AED modules. These are separate models, but have been set up to run side by side. They are configured using the text file aed2.nml.
The above simulation was only plotting temperature and salinity and these plots were configured in plots.nml. Open plots.nml to see how you can customise the graphs. There are many other variables we may be interested in plotting when we run the watter quality model such as oxygen, nutrients, and algae. A pre-configured file called plots_aed2.nml has been created. To use this file, rename it to plots.nml (you’ll first need to rename the existing plots.nml) and run the model. You should now see more outputs for the lake simulation.
Let’s also now add water quality variables to the specific depth output files (i.e. WQ_5.csv). To do so we must edit the output section of the glm3.nml file, by adding the variables as extra columns to the .csv file that were configured above.
In plots.nml the required variables are all listed, make sure the number of plots is high enough to include them all.
| Description | Variable Name |
|---|---|
| Passive tracer |
TRC_tr1
|
| Water age |
TRC_age
|
| Suspended solids |
NCS_ss1
|
| Dissolved oxygen |
OXY_oxy
|
| Dissolved inorganic carbon |
CAR_dic
|
| pH |
CAR_pH
|
| Methane (dissolved) |
CAR_ch4
|
| Methane (bubbles) |
CAR_ch4_bub
|
| Reactive Silica |
SIL_rsi
|
| Ammonium |
NIT_amm
|
| Nitrate/Nitrite |
NIT_nit
|
| Phosphate |
PHS_frp
|
| Particulate inorganic phosphorus |
PHS_frp_ads
|
| Dissolved organic carbon |
OGM_doc
|
| Particualte organic carbon |
OGM_poc
|
| Dissolved organic nitrogen |
OGM_don
|
| Dissolved organic nitrogen |
OGM_pon
|
| Particualte organic phosphorus |
OGM_dop
|
| Dissolved organic phsophorus |
OGM_pop
|
| Green algae |
PHY_green
|
| Green algae |
PHY_green_IN
|
| Green algae |
PHY_green_IP
|
| Diatoms |
PHY_diatom
|
| Diatoms |
PHY_diatom_IN
|
| Diatoms |
PHY_diatom_IP
|
| Cyrptophytes |
PHY_crypto
|
| Cyrptophytes |
PHY_crypto_IN
|
| Cyrptophytes |
PHY_crypto_IP
|
| Zooplankton |
ZOO_zoo01
|
Now run the model as above by double clicking the glm.bat.
- Create a well formatted, interesting graph of variables of your choice (for example, you may like to consider all of the phytoplankton groups, all of the nitrogen variables, or create sums of these to generate a NPZD model) to see how they interact and change over time. If any variables are much bigger or smaller than the others, then use two y axes or multiple plots.
- How would these concentrations change if the air temperature increased by 2°C?
(You will need to edit the
met_hourly.csvfile). Make sure to save with correct date format, taking note these met measurements are recorded hourly, so YYYY-MM-DD hh:mm. A pre-made version of this file can be found in bcs which is calledmet_1997_2004_airT2.csv. To use this file, updatemeteo_fl = 'bcs/met_1997_2004.csv'in the&meteorologysection ofglm3.nml.
!-------------------------------------------------------------------------------
! meteorology
!-------------------------------------------------------------------------------
&meteorology
met_sw = .true.
lw_type = 'LW_IN'
rain_sw = .false.
atm_stab = 0
fetch_mode = 0
!rad_mode = 1
albedo_mode = 1
cloud_mode = 4
!-- BC file details
subdaily = .true.
meteo_fl = 'bcs/met_1997_2004.csv' <------- Update this!
wind_factor = 0.9
lw_factor = 1.0
lw_offset = 0.0
!-- Parameters
ce = 0.0013
ch = 0.0013
cd = 0.0013
!catchrain = .true.
!rain_threshold = 0.001
!runoff_coef = 0.0
/- What happens if we change the water clarity? Increase and decrease from the original value to see the response
Kw (range 0-1) in the glm.nml file.
Assessing Error & Sensitivity
- Calculate the mean absolute error in the simulation using the observed temperature data (
ObservedTempData.xlsxfrom the data folder in Kinneret97) and the simulated temperature data (the simulated temperature data is from the surface layer inlake.csv).
- Calculate the sensitivity of the modelled temperature to changes in water clarity (the light extinction coefficient, \(K_w\)) and wind speed (
wind_factor). These can be found inglm.nml. Try increasing and decreasing the default parameter value by 0.2 and see how much the output changes.
- Assess how sensitive the phytoplankton biomass is to water clarity:
| GLM results | Water clarity Decrease 0.37 | Water clarity Original 0.57 | Water clarity Increase 0.77 |
|---|---|---|---|
| Average WQ_35 temperature | |||
| Average WQ_35 phytoplankton (green, crypto, diatom) biomass |
- Assess how sensitive the phytoplankton biomass is to wind speed:
| GLM results | Wind Speed Decrease 0.37 | Wind Speed Original 0.57 | Wind Speed Increase 0.77 |
|---|---|---|---|
| Average WQ_35 temperature | |||
| Average WQ_35 phytoplankton (green, crypto, diatom) biomass |
Submission
Submit properly formatted graphs and tables of the following sections of the lab:
Lake Stratification:
- Hand drawn diagram of the lake and any variables you used
- Plots of the water balance
- Plots of the heat fluxes
- Plots of the temperature at different layers
- Plots of the shallower lake
The Ecosystem Model:
- Hand drawn diagram of the lake with any variables you used
- Interesting plot of the selected variables
- Any results of the higher air temperature and changed water clarity
Assessing Error & Sensitivity:
- Temperature error assessment
- Temperature and phytoplankton sensitivity assessment
- Tables for submission
These are to be uploaded in a word doc or PDF format. No screenshots of figures from Excel/Excel spreadsheets to be uploaded
General professional formatting guidelines:
- All figures are to have adequate captions explaining them
- For graphs, figure captions go below the plot
- For tables, the caption goes above the table
- Make sure figures and their text size is readable
Excel hints:
- When there is a caption for a plot, you remove the title
- Remove the plot border and gridlines
- Make sure both axes have visible lines and tick marks
- Units need to be noted properly with the axis label - ‘Temperature (°C)’
- Round numbers to be reasonable