3 Organisation of the Library & Module Structure
Author: Matthew R. Hipsey
3.1 Library Design
The AED modelling library is organised as a series of “modules” that can be connected or “linked” through specific variable dependencies. By linking modules users can simulate most aquatic biogeochemical processes relevant to nutrient cycling, oxygen dynamics, harmful algal blooms, sediment/soil biogeochemistry, vegetation, pathogens etc. Each module can work alone or combined with other modules, depending on the complexity of targeted system.
A wide array of modules are available with some operational only in water, some operational where no water is present, and some can operate under both dry and wet conditions.
The current AED software is divided into 5 separate code-libraries: water, benthic, demo, dev, and riparian. Prior to the 2020 release, the model was distributed as two libraries: the first was the core AED2 library (libaed2) and the second is the advanced modules which are bundled together as AED2+ (libaed2-plus).
Currently there are 8 core modules in AED, and 12 advanced modules in AED+. For routine water quality assessments of lakes or estuaries, AED will suit most applications. For advanced users and researchers seeking to extend their simulation abilities, then AED+ includes numerous advanced options.
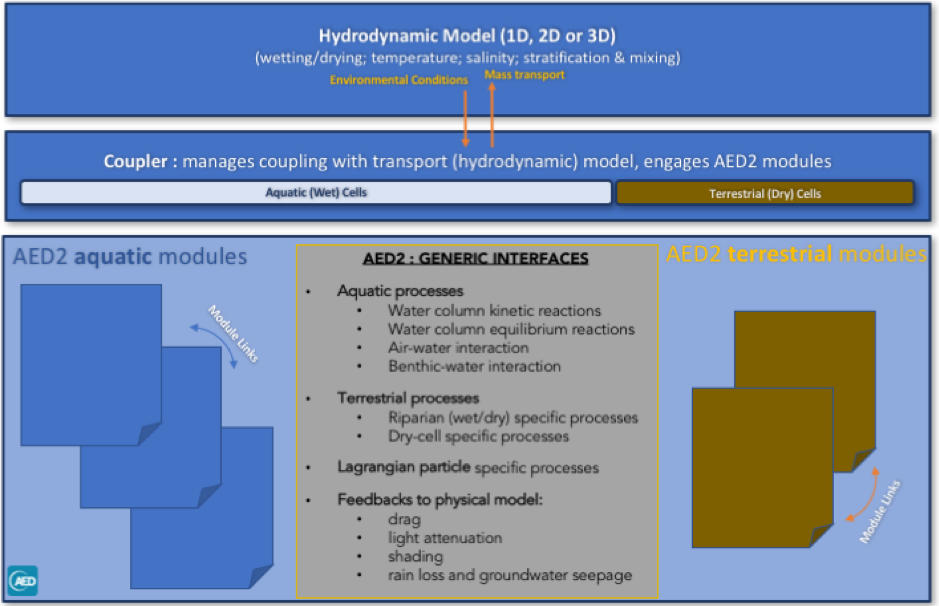
3.2 Model Structure
The philosophy of the AED/AED2 library is that biogeochemical and ecological modules have a standard class structure. State variables defined within each module are subject to transport and mass conservation, as managed by the scalar transport facility of the host hydrodynamic model (see Section 4).
Any AED module can have the ability to simulate a range of physical, chemical and biological processes, that can be generally classified as belonging to:
- Feedback of chemical or biological attributes to physical properties of water (light extinction, drag, density)
- Water column kinetic (time-varying) chemical / biological transformations (e.g., denitrification or algal growth)
- Water column equilibrium (instantaneous) chemical transformations (e.g., PO4 adsorption)
- Biogeochemical transformations in the sediment or biological changes in the benthos
- Vertical sedimentation or migration
- Fluxes across the air-water interface
- Fluxes across the sediment-water interface
- Fluxes across the terrestrial-water interface (riparian interactions)
- Ecohydrological dynamics and biogeochemical transformations in the exposed (dry) cells
- Feedbacks of soil and vegetation dynamics onto lake hydrodynamics and water balance
A general module structure in the figure above. Specific methods available within a module include those shown in the table. Note that not all methods must be used in modules; developers must choose which methods are relevant to a specific module. The only exception is “define”, which is compulsory for all modules to configure the chosen state variables, diagnostic variables, and set the necessary parameters and options.
3.3 Module Summary
A high level overview of the simulatable modules are include in Table 3.1 below.
| Module | Description | Relevant Variables |
|---|---|---|
| TRC: Tracers, Suspended Soilds and Retention Time | Modellers can use the aed_tracer to simulate a dissolved or particulate tracer (subject to transport processes only), or this can be optionally configured to account for decay, sedimentation and/or resuspension. This module also include an option to simulate water “retention time” where the water age increments once enters into the waterbody | C,\(t_{ret}\) |
| OXY: Dissolved Oxygen | Dissolved oxygen (DO) dynamics are able to be simulated, accounting for atmospheric exchange and sediment oxygen demand, and through links to other modules will account for microbial use during organic matter mineralisation and nitrification, photosynthetic oxygen production and respiratory oxygen consumption, and respiration by other optional biotic components | O2 |
| NUT: Inorganic Nutrients | Modules exist for simulation of inorganic nutrients including phosphorus, nitrogen and silica. These modules provide basic nutrient cycling functionality and are designed to be linked with other modules (eg OGM, PHY) in order to provide a more comprehensive depiction of nutrient cycling. | |
| OGM: Organic Matter | Organic matter variables cover the C, N & P stored in the dissolved and particulate organic matter pools. This module optionally also supports depiction of “labile” vs “refractory” fractions of organic matter, including the breakdown and hydrolysis process, photo-degradation and mineralisation. | DOM, POM |
| PHY: Phytoplankton | Highly customisable phytoplankton module for simulating change in algae, cyano-bacteria and chl-a, including phytoplankton production/respiration, nutrient uptake, excretion, vertical movement (eg buoyancy control), and grazing effects. Benthic phytoplankton may also be optionally configured. | |
| ZOO: Zooplankton | Simulates different size classes of zooplankton, accounting for carbon and nutrient assimilation from grazing, carbon loss via respiration, excretion of DOM, faecal pellet production, mortality, and predation by larger organisms. | |
| TOT: Totals | A summary module, allowing users to “sum-up” component variables from other modules into a total, for example, to compute TN, TP or TSS. | |
| SDF: Sediment Flux | An interface module designed to provide spatially variable sediment flux settings to key modules (e.g., OXY, OGM, NUT), and/or link these variables to the dynamic sediment biogeochemistry model (SDG). |
| Module | Description | Relevant Variables |
|---|---|---|
| MAC: Benthic macrophytes | Simulates benthic habitat and/or growth of macrophytes such as seagrasses in specified sediment zones. | NA |
| MAG: Macroalgae | Simulates benthic macroalgal growth and sloughing/detachment of macroalgae and its subsequent redistribution within the domain. | NA |
| BIV: Bivalves | A model of one or more groups of benthic filter feeders, able to assimilate C, N and P and recycle filter material back to the water column and sediment. | NA |
| HAB: Habitat Quality | A high level module that returns habitat quality metrics based on underlying water condition attributes, for example, for seagrass or crab habitat suitability at different life stages. | NA |